## Genome assembly Lab 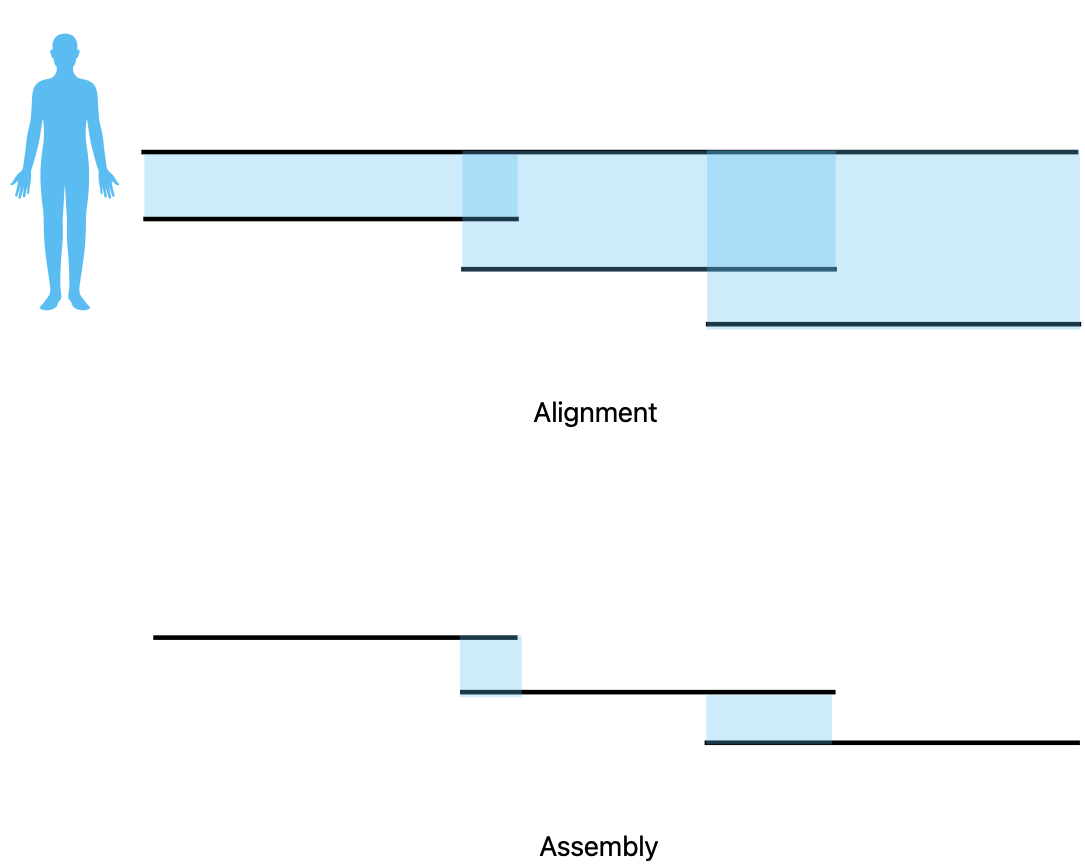 <!-- .element height="80%" width="80%" --> --- ## NCBI assembly model 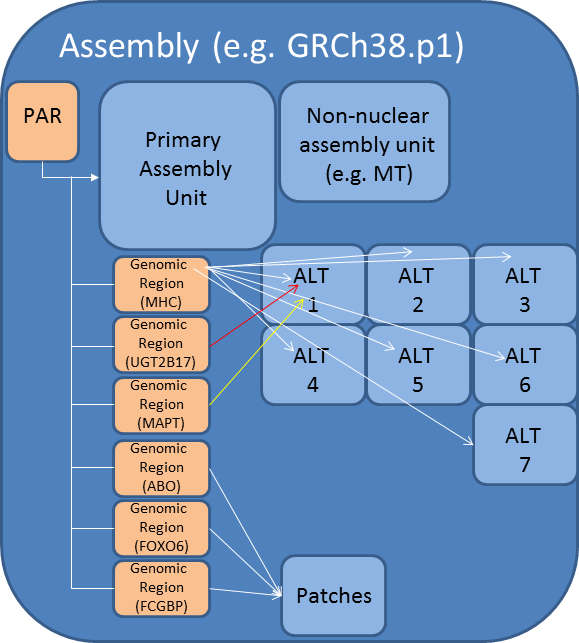 <small>Source: [NCBI Genome Assembly Model](https://www.ncbi.nlm.nih.gov/assembly/model/)</small> Notes: Sequence updates that are released outside of the major assembly cycle are called patches. There are two types of patches: Fix patch: This patch is made in a region where the Tiling Path File (TPF) will change in the next major assembly update. These scaffolds will be withdrawn at the next major assembly update, the accessions will be made secondary to the chromosome and the sequence will be incorporated into the Primary Assembly TPF. Novel patch: These represent new alternate loci. At the next major assembly update, these sequences will be moved to the appropriate assembly-unit and the accession will remain stable. A sequence that provides an alternate representation of a locus found in the haploid assembly is called a alt loci. PAR Pseudo-autosomal region. A region found on the X and Y chromosomes of mammals that allows recombination between the sex chromosomes --- ## Which human genome should I use? Potential issues * Inclusion of ALT contigs * Inclusion of multi-placed sequences (e.g., PAR) * Chromosome names * Unplaced and unlocalized contigs [Heng Li: Which human reference genome to use](https://lh3.github.io/2017/11/13/which-human-reference-genome-to-use) --- ## Pangenomes 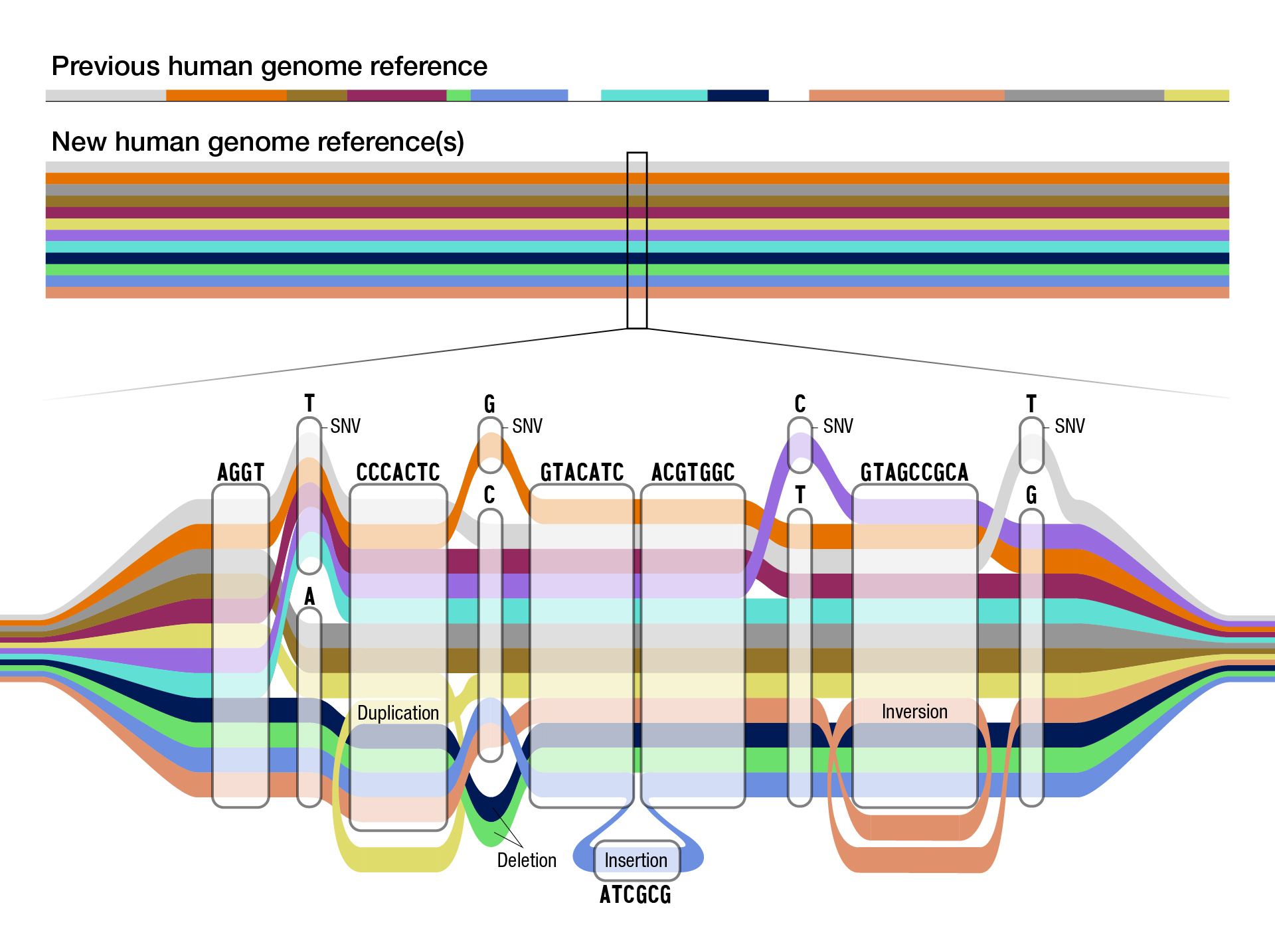 <!-- .element height="80%" width="80%" --> <small>[Scientists release a new human “pangenome” reference](https://www.genome.gov/news/news-release/scientists-release-a-new-human-pangenome-reference) </small> --- ## Pangenomes  <small>[PacBio](https://www.pacb.com/blog/the-hifi-difference-enabling-the-human-pangenome-reference) </small> --- ## Assignment * [https://github.com/cphg/genome_assembler](https://github.com/cphg/genome_assembler) * Implement an algorithm to simplify overlap graph by iteratively removing transitively-inferrible edges * implement the `simplify` function in `remove_transitive_edges.py` * test your work `python3 test_remove_transitive_edges` * Skeleton code is provided in python * A simple graph data structure has already been implemented for you. Use it. * Simple test cases are also provided * Clone the repo before making changes --- ## Shortest Common Superstring Given set of strings $S$ find $SCS(S)$: shortest string containing the strings in $S$ as substrings  --- ## Idea 1: Enumerate all orders  --- ## Idea 1: Enumerate all orders  Try all possible orderings and pick shortest. Note: If S contains n strings, then how many orderings (n!). The SCS problem is NP-hard. Non-deterministic polynomial time hard problems contain the most complex problems in computer science. They are not only hard to solve but are hard to verify as well. In fact, some of these problems aren’t even decidable. --- ## Idea 2: Choose order prioritizing similarity  --- ## Idea 2: Choose order prioritizing similarity  Note: We keep on doing this, and we will have AAABBBA or AAABBAB as the result. --- ## Shortest common superstring  Note: The answer might be different if we try an equally reasonable order. For example, if we use ABB,BBA in the second step, then we get a superstring of length 9 --- ## Shortest common superstring The greedy algorithm is a good approximation; i.e. the superstring yielded by the greedy algorithm won’t be more than ~2.5 times longer than true SCS (see Gusfield, Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology, 16.17.1) Note: Greedy algorithm is not guaranteed to choose overlaps yielding SCS --- ## Programming Problem for fun * [https://github.com/cphg/genome_assembler](https://github.com/cphg/genome_assembler) * Skeleton code is provided in python * Simple test cases are also provided * Clone the repo and then make changes * Implement parts of the Shortest common superstring problem * implement the `calculate_scs` function in `shortest_common_superstring.py` * test your work `python3 test_shortest_common_superstring` ---