Data Visualization with ggplot2
Last updated on 2025-11-11 | Edit this page
Estimated time: 90 minutes
Overview
Questions
- What is ggplot2?
- What is mapping, and what is aesthetics?
- What is the process of creating a publication-quality plots with ggplot in R?
Objectives
- Describe the role of data, aesthetics, and geoms in ggplot functions.
- Choose the correct aesthetics and alter the geom parameters for a scatter plot, histogram, or box plot.
- Layer multiple geometries in a single plot.
- Customize plot scales, titles, themes, and fonts.
- Apply a facet to a plot.
- Apply additional ggplot2-compatible plotting libraries.
- Save a ggplot to a file.
- List several resources for getting help with ggplot.
- List several resources for creating informative scientific plots.
Introduction to ggplot2

ggplot2 is a plotting package, part of
the tidyverse, that makes it simple to create complex plots from data in
a data frame. It provides a more programmatic interface for specifying
what variables to plot, how they are displayed, and general visual
properties. Therefore, we only need minimal changes if the underlying
data change or if we decide to change from a bar plot to a scatter plot.
This helps in creating publication-quality plots with minimal amounts of
adjustments and tweaking.
The gg in “ggplot” stands for “Grammar of Graphics,” which is an elegant yet powerful way to describe the making of scientific plots. In short, the grammar of graphics breaks down every plot into a few components, namely, a dataset, a set of geoms (visual marks that represent the data points), and a coordinate system. You can imagine this is a grammar that gives unique names to each component appearing in a plot and conveys specific information about data. With ggplot, graphics are built step by step by adding new elements.
The idea of mapping is crucial in
ggplot. One familiar example is to map the
value of one variable in a dataset to \(x\) and the other to \(y\). However, we often encounter datasets
that include multiple (more than two) variables. In this case,
ggplot allows you to map those other variables
to visual marks such as color and
shape (aesthetics or
aes). One thing you may want to remember is the difference
between discrete and continuous
variables. Some aesthetics, such as the shape of dots, do not accept
continuous variables. If forced to do so, R will give an error. This is
easy to understand; we cannot create a continuum of shapes for a
variable, unlike, say, color.
Tip: when having doubts about whether a variable is
continuous
or discrete, a quick way to check is to use the summary()
function. Continuous variables have descriptive statistics but not the
discrete variables.
Loading packages
Let’s load the ggplot2 package:
R
library(ggplot2)
We will also use some of the other tidyverse packages we used in the last episode, so we need to load them as well.
R
library(readr)
library(dplyr)
OUTPUT
Attaching package: 'dplyr'OUTPUT
The following objects are masked from 'package:stats':
filter, lagOUTPUT
The following objects are masked from 'package:base':
intersect, setdiff, setequal, unionAs we can see from above output ggplot2
has been already loaded along with other packages as part of the
tidyverse framework.
Note on saving plots/graphics
R can be used to create complex plots from data, and we typically
save them in files so the plots can be included in reports,
presentations, and manuscripts. Let’s consider an example of a scatter
plot and see how we would save it and then view it on the HPC.
mtcars is a data.frame that includes data extracted from
the 1974 Motor Trend US magazine, and comprises fuel consumption and 10
aspects of automobile design and performance for 32 automobiles. Let’s
check if there is a correlation between Miles/(US) gallon and Weight
(lb/1000) of those cars using visual inspection
R
x <- mtcars$mpg
y <- mtcars$wt
pdf("mpg_vs_wt.pdf")
plot(x, y, xlab="mpg", ylab="wt")
dev.off()
OUTPUT
png
2 The above code does the following step by step:
-
x <- mtcars$mpgextracts the miles per gallon column into x. -
y <- mtcars$wtextracts the weight (1000 lbs) column into y. -
pdf("mpg_vs_wt.pdf")opens a PDF graphics device, so all plots are written to mpg_vs_wt.pdf. -
plot(x, y, xlab="mpg", ylab="wt")creates a scatterplot with custom axis labels. -
dev.off()closes the PDF device, finalizing the file.
To view the plot, go to https://ood.hpc.virginia.edu/pun/sys/dashboard,
and log in with your credentials. This will bring you to a unified
dashboard that provides access to your HPC resources. From the top menu,
open the Files tab and select Home Directory
to launch a file explorer. Navigate to the day2 folder,
where you should find a file named mpg_vs_wt.pdf. Clicking
on this file will open the plot directly in your browser. As the plot
suggests, there might be a negative correlation between the two
variables.
So, whenever you create a plot, save it as a PDF with a clear, descriptive name. You can then view the file through https://ood.hpc.virginia.edu/pun/sys/dashboard.
Loading the dataset
R
variants <- read.csv("https://raw.githubusercontent.com/datacarpentry/genomics-r-intro/main/episodes/data/combined_tidy_vcf.csv")
Explore the structure (types of columns and number of rows)
of the dataset using dplyr’s glimpse()
(for more info, see the Data
Wrangling and Analyses with Tidyverse episode)
R
glimpse(variants) # Show a snapshot of the rows and columns
OUTPUT
Rows: 801
Columns: 29
$ sample_id <chr> "SRR2584863", "SRR2584863", "SRR2584863", "SRR2584863", …
$ CHROM <chr> "CP000819.1", "CP000819.1", "CP000819.1", "CP000819.1", …
$ POS <int> 9972, 263235, 281923, 433359, 473901, 648692, 1331794, 1…
$ ID <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ REF <chr> "T", "G", "G", "CTTTTTTT", "CCGC", "C", "C", "G", "ACAGC…
$ ALT <chr> "G", "T", "T", "CTTTTTTTT", "CCGCGC", "T", "A", "A", "AC…
$ QUAL <dbl> 91.0000, 85.0000, 217.0000, 64.0000, 228.0000, 210.0000,…
$ FILTER <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ INDEL <lgl> FALSE, FALSE, FALSE, TRUE, TRUE, FALSE, FALSE, FALSE, TR…
$ IDV <int> NA, NA, NA, 12, 9, NA, NA, NA, 2, 7, NA, NA, NA, NA, NA,…
$ IMF <dbl> NA, NA, NA, 1.000000, 0.900000, NA, NA, NA, 0.666667, 1.…
$ DP <int> 4, 6, 10, 12, 10, 10, 8, 11, 3, 7, 9, 20, 12, 19, 15, 10…
$ VDB <dbl> 0.0257451, 0.0961330, 0.7740830, 0.4777040, 0.6595050, 0…
$ RPB <dbl> NA, 1.000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.900802, …
$ MQB <dbl> NA, 1.0000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.1501340…
$ BQB <dbl> NA, 1.000000, NA, NA, NA, NA, NA, NA, NA, NA, 0.750668, …
$ MQSB <dbl> NA, NA, 0.974597, 1.000000, 0.916482, 0.916482, 0.900802…
$ SGB <dbl> -0.556411, -0.590765, -0.662043, -0.676189, -0.662043, -…
$ MQ0F <dbl> 0.000000, 0.166667, 0.000000, 0.000000, 0.000000, 0.0000…
$ ICB <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ HOB <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ AC <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ AN <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ DP4 <chr> "0,0,0,4", "0,1,0,5", "0,0,4,5", "0,1,3,8", "1,0,2,7", "…
$ MQ <int> 60, 33, 60, 60, 60, 60, 60, 60, 60, 60, 25, 60, 10, 60, …
$ Indiv <chr> "/home/dcuser/dc_workshop/results/bam/SRR2584863.aligned…
$ gt_PL <chr> "121,0", "112,0", "247,0", "91,0", "255,0", "240,0", "20…
$ gt_GT <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ gt_GT_alleles <chr> "G", "T", "T", "CTTTTTTTT", "CCGCGC", "T", "A", "A", "AC…Alternatively, we can display the first a few rows (vertically) of
the table using head():
R
head(variants)
OUTPUT
sample_id CHROM POS ID REF ALT QUAL FILTER INDEL IDV IMF
1 SRR2584863 CP000819.1 9972 NA T G 91 NA FALSE NA NA
2 SRR2584863 CP000819.1 263235 NA G T 85 NA FALSE NA NA
3 SRR2584863 CP000819.1 281923 NA G T 217 NA FALSE NA NA
4 SRR2584863 CP000819.1 433359 NA CTTTTTTT CTTTTTTTT 64 NA TRUE 12 1.0
5 SRR2584863 CP000819.1 473901 NA CCGC CCGCGC 228 NA TRUE 9 0.9
6 SRR2584863 CP000819.1 648692 NA C T 210 NA FALSE NA NA
DP VDB RPB MQB BQB MQSB SGB MQ0F ICB HOB AC AN DP4 MQ
1 4 0.0257451 NA NA NA NA -0.556411 0.000000 NA NA 1 1 0,0,0,4 60
2 6 0.0961330 1 1 1 NA -0.590765 0.166667 NA NA 1 1 0,1,0,5 33
3 10 0.7740830 NA NA NA 0.974597 -0.662043 0.000000 NA NA 1 1 0,0,4,5 60
4 12 0.4777040 NA NA NA 1.000000 -0.676189 0.000000 NA NA 1 1 0,1,3,8 60
5 10 0.6595050 NA NA NA 0.916482 -0.662043 0.000000 NA NA 1 1 1,0,2,7 60
6 10 0.2680140 NA NA NA 0.916482 -0.670168 0.000000 NA NA 1 1 0,0,7,3 60
Indiv gt_PL
1 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 121,0
2 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 112,0
3 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 247,0
4 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 91,0
5 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 255,0
6 /home/dcuser/dc_workshop/results/bam/SRR2584863.aligned.sorted.bam 240,0
gt_GT gt_GT_alleles
1 1 G
2 1 T
3 1 T
4 1 CTTTTTTTT
5 1 CCGCGC
6 1 Tggplot2 functions like data in the
long format, i.e., a column for every dimension
(variable), and a row for every observation. Well-structured data will
save you time when making figures with
ggplot2
ggplot2 graphics are built step-by-step
by adding new elements. Adding layers in this fashion allows for
extensive flexibility and customization of plots, and more equally
important the readability of the code.
To build a ggplot, we will use the following basic template that can be used for different types of plots:
- use the
ggplot()function and bind the plot to a specific data frame using thedataargument. Here, we save it in a file namedscatterplot.pdfwhich you can view through https://ood.hpc.virginia.edu/pun/sys/dashboard.
R
pdf("scatterplot.pdf")
ggplot(data = variants)
dev.off()
- define a mapping (using the aesthetic (
aes) function), by selecting the variables to be plotted and specifying how to present them in the graph, e.g. as x and y positions or characteristics such as size, shape, color, etc. We will overwrite the previous plot, but feel free to save this plot in a different file.
R
pdf("scatterplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP))
dev.off()
- add ‘geoms’ – graphical representations of the data in the plot
(points, lines, bars).
ggplot2offers many different geoms; we will use some common ones today, including:-
geom_point()for scatter plots, dot plots, etc. -
geom_boxplot()for, well, boxplots! -
geom_line()for trend lines, time series, etc.
-
To add a geom to the plot use the + operator. Because we
have two continuous variables, let’s use geom_point()
(i.e., a scatter plot) first:
R
pdf("scatterplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP)) +
geom_point()
dev.off()
OUTPUT
png
2 The + in the ggplot2
package is particularly useful because it allows you to modify existing
ggplot objects. This means you can easily set up plot
templates and conveniently explore different types of plots, so the
above plot can also be generated with code like this:
R
# Assign plot to a variable
coverage_plot <- ggplot(data = variants, aes(x = POS, y = DP))
# Draw the plot
pdf("scatteplot.pdf")
coverage_plot +
geom_point()
dev.off()
Notes
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e., these are universal plot settings). This includes the x- and y-axis mapping you set up inaes(). - You can also specify mappings for a given geom independently of the
mappings defined globally in the
ggplot()function. - The
+sign used to add new layers must be placed at the end of the line containing the previous layer. If, instead, the+sign is added at the beginning of the line containing the new layer,ggplot2will not add the new layer and will return an error message.
R
# This is the correct syntax for adding layers
coverage_plot +
geom_point()
# This will not add the new layer and will return an error message
coverage_plot
+ geom_point()
Building your plots iteratively
Building plots with ggplot2 is
typically an iterative process. We start by defining the dataset we’ll
use, lay out the axes, and choose a geom:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP)) +
geom_point()
dev.off()
OUTPUT
png
2 Then, we start modifying this plot to extract more information from
it. For instance, we can add transparency (alpha) to avoid
over-plotting:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP)) +
geom_point(alpha = 0.5)
dev.off()
OUTPUT
png
2 We can also add colors for all the points:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP)) +
geom_point(alpha = 0.5, color = "blue")
dev.off()
OUTPUT
png
2 Or to color each species in the plot differently, you could use a
vector as an input to the argument color.
ggplot2 will provide a different color
corresponding to different values in the vector. Here is an example
where we color with sample_id:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP, color = sample_id)) +
geom_point(alpha = 0.5)
dev.off()
OUTPUT
png
2 Notice that we can change the geom layer and colors will be still
determined by sample_id
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP, color = sample_id)) +
geom_line(alpha = 0.5)
dev.off()
OUTPUT
png
2 To make our plot more readable, we can add axis labels:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP, color = sample_id)) +
geom_point(alpha = 0.5) +
labs(x = "Base Pair Position",
y = "Read Depth (DP)")
dev.off()
OUTPUT
png
2 To add a main title to the plot, we use the title
argument for the labs() function:
R
pdf("scatteplot.pdf")
ggplot(data = variants, aes(x = POS, y = DP, color = sample_id)) +
geom_point(alpha = 0.5) +
labs(x = "Base Pair Position",
y = "Read Depth (DP)",
title = "Read Depth vs. Position")
dev.off()
OUTPUT
png
2 Challenge
Use what you just learned to create a scatter plot of mapping quality
(MQ) over position (POS) with the samples
showing in different colors. Make sure to give your plot relevant axis
labels. Save the output to a file named challenge.pdf
R
pdf('challenge.pdf')
ggplot(data = variants, aes(x = POS, y = MQ, color = sample_id)) +
geom_point() +
labs(x = "Base Pair Position",
y = "Mapping Quality (MQ)")
dev.off()
OUTPUT
png
2 To further customize the plot, we can change the default font format. We do not include the commands to save the plot from here on, but you should save and view the plots.
R
ggplot(data = variants, aes(x = POS, y = DP, color = sample_id)) +
geom_point(alpha = 0.5) +
labs(x = "Base Pair Position",
y = "Read Depth (DP)",
title = "Read Depth vs. Position") +
theme(text = element_text(family = "Bookman"))
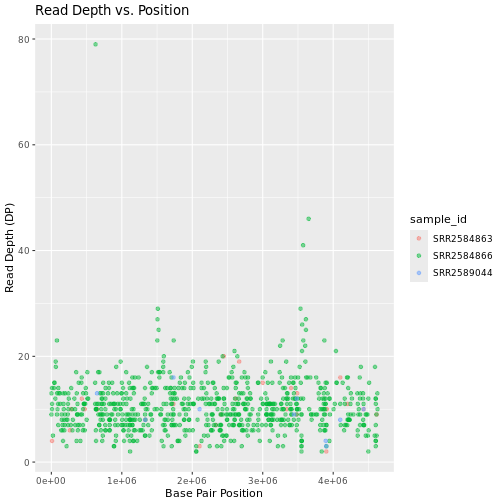
Faceting
ggplot2 has a special technique called
faceting that allows the user to split one plot into multiple
plots (panels) based on a factor (variable) included in the dataset. We
will use it to split our mapping quality plot into three panels, one for
each sample.
R
ggplot(data = variants, aes(x = POS, y = MQ, color = sample_id)) +
geom_point() +
labs(x = "Base Pair Position",
y = "Mapping Quality (MQ)") +
facet_grid(~ sample_id)
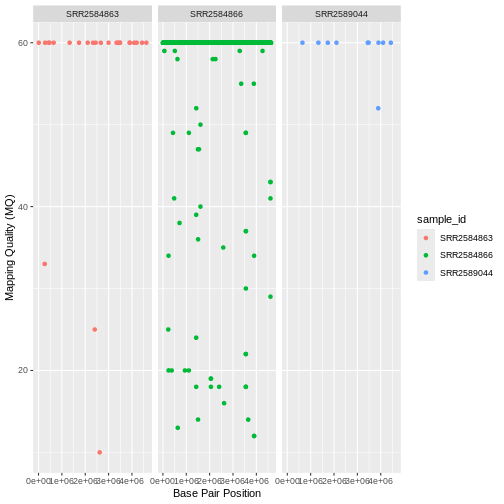
This looks okay, but it would be easier to read if the plot facets
were stacked vertically rather than horizontally. The
facet_grid geometry allows you to explicitly specify how
you want your plots to be arranged via formula notation
(rows ~ columns; the dot (.) indicates every
other variable in the data i.e., no faceting on that side of the
formula).
R
ggplot(data = variants, aes(x = POS, y = MQ, color = sample_id)) +
geom_point() +
labs(x = "Base Pair Position",
y = "Mapping Quality (MQ)") +
facet_grid(sample_id ~ .)
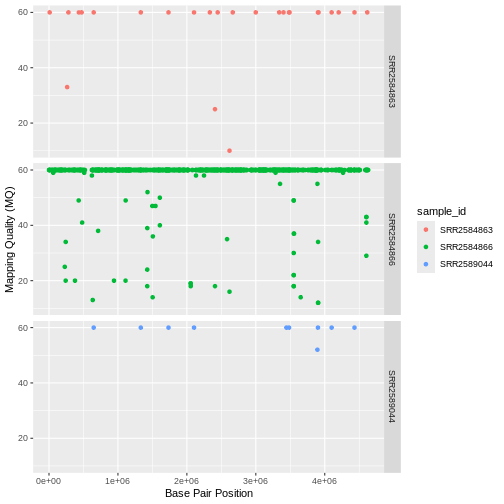
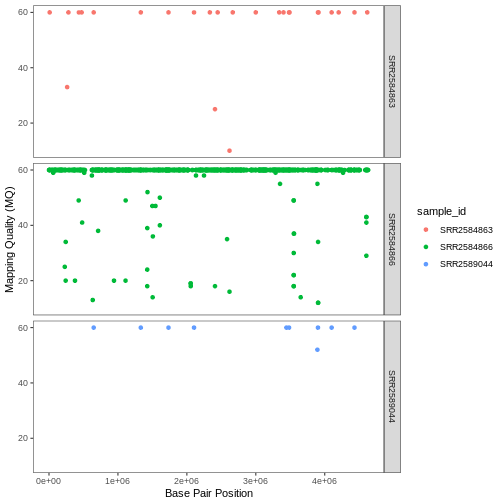
Usually plots with white background look more readable when printed.
We can set the background to white using the function theme_bw().
Additionally, you can remove the grid:
R
ggplot(data = variants, aes(x = POS, y = MQ, color = sample_id)) +
geom_point() +
labs(x = "Base Pair Position",
y = "Mapping Quality (MQ)") +
facet_grid(sample_id ~ .) +
theme_bw() +
theme(panel.grid = element_blank())
Challenge
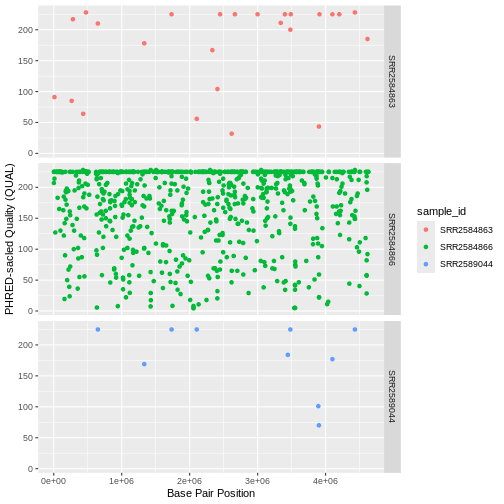
Use what you just learned to create a scatter plot of PHRED scaled
quality (QUAL) over position (POS) with the
samples showing in different colors. Make sure to give your plot
relevant axis labels.
R
ggplot(data = variants, aes(x = POS, y = QUAL, color = sample_id)) +
geom_point() +
labs(x = "Base Pair Position",
y = "PHRED-sacled Quality (QUAL)") +
facet_grid(sample_id ~ .)
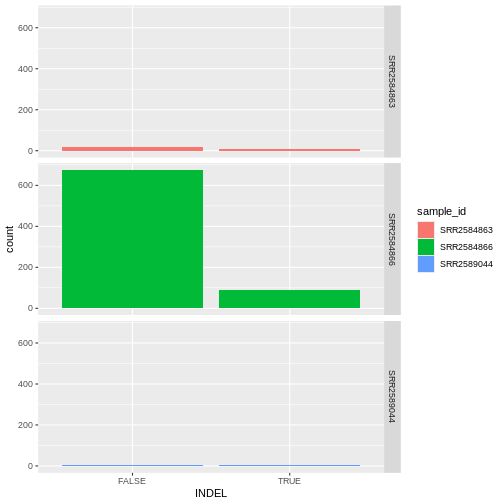
Barplots
We can create barplots using the geom_bar
geom. Let’s make a barplot showing the number of variants for each
sample that are indels.
R
ggplot(data = variants, aes(x = INDEL, fill = sample_id)) +
geom_bar() +
facet_grid(sample_id ~ .)
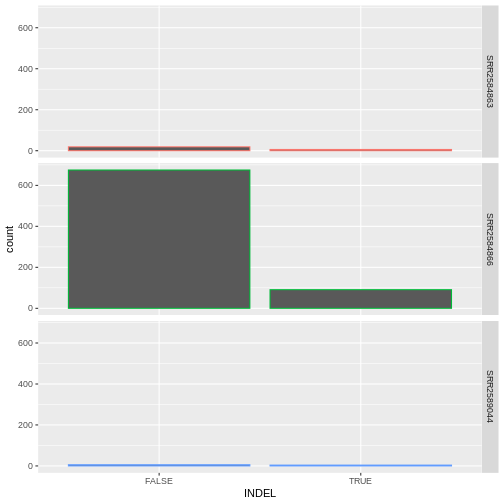
Challenge
Since we already have the sample_id labels on the individual plot
facets, we don’t need the legend. Use the help file for
geom_bar and any other online resources you want to use to
remove the legend from the plot.
R
ggplot(data = variants, aes(x = INDEL, color = sample_id)) +
geom_bar(show.legend = F) +
facet_grid(sample_id ~ .)
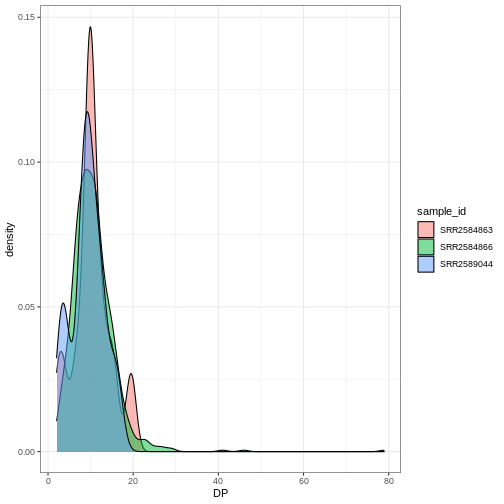
Density
We can create density plots using the geom_density
geom that shows the distribution of of a variable in the dataset. Let’s
plot the distribution of DP
R
ggplot(data = variants, aes(x = DP)) +
geom_density()
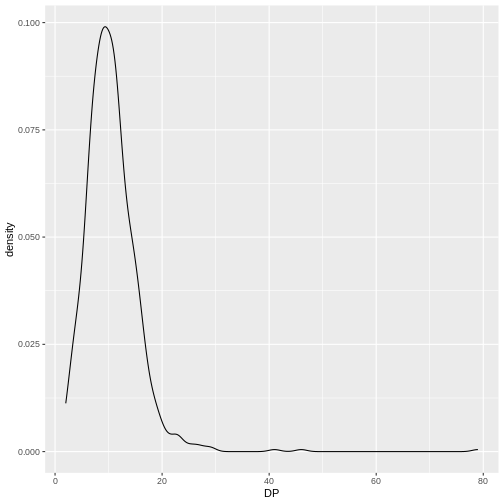
This plot tells us that the most of frequent DP (read
depth) for the variants is about 10 reads.
Challenge
Use geom_density
to plot the distribution of DP with a different fill for
each sample. Use a white background for the plot.
R
ggplot(data = variants, aes(x = DP, fill = sample_id)) +
geom_density(alpha = 0.5) +
theme_bw()
ggplot2 themes
In addition to theme_bw(),
which changes the plot background to white,
ggplot2 comes with several other themes
which can be useful to quickly change the look of your visualization.
The complete list of themes is available at https://ggplot2.tidyverse.org/reference/ggtheme.html.
theme_minimal() and theme_light() are popular,
and theme_void() can be useful as a starting point to
create a new hand-crafted theme.
The ggthemes
package provides a wide variety of options (including Microsoft Excel,
old
and new).
The ggplot2
extensions website provides a list of packages that extend the
capabilities of ggplot2, including
additional themes.
Challenge
With all of this information in hand, please take another five
minutes to either improve one of the plots generated in this exercise or
create a beautiful graph of your own. Use the RStudio ggplot2
cheat sheet for inspiration. Here are some ideas:
- See if you can change the size or shape of the plotting symbol.
- Can you find a way to change the name of the legend? What about its labels?
- Try using a different color palette (see the Cookbook for R).
More ggplot2 Plots
ggplot2 offers many more informative
and beautiful plots (geoms) of interest for biologists
(although not covered in this lesson) that are worth exploring, such
as
-
geom_tile(), for heatmaps, -
geom_jitter(), for strip charts, and -
geom_violin(), for violin plots
Resources
- ggplot2: Elegant Graphics for Data Analysis (online version)
- The Grammar of Graphics (Statistics and Computing)
- Data Visualization: A Practical Introduction (online version)
- The R Graph Gallery (the book)
- ggplot2 is a powerful tool for high-quality plots
- ggplot2 provides a flexible and readable grammar to build plots
